The central dogma of molecular biology¶
(over)simplified: “DNA makes RNA, RNA makes proteins, proteins make us.”

- Transcription
- Splicing
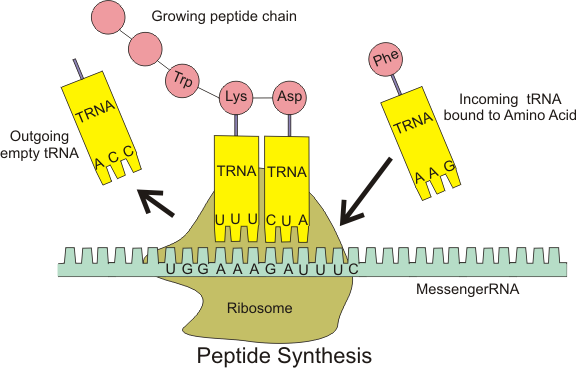
- Translation
Genes active and their transcription rates and are different for different cells.
from IPython.display import YouTubeVideo
YouTubeVideo('2BwWavExcFI', width=1024, height=576)
There are other types of RNA besides messenger RNA (mRNA): transfer RNA (tRNA), ribosomal RNA (rRNA), small and long non coding RNA (ncRNA).
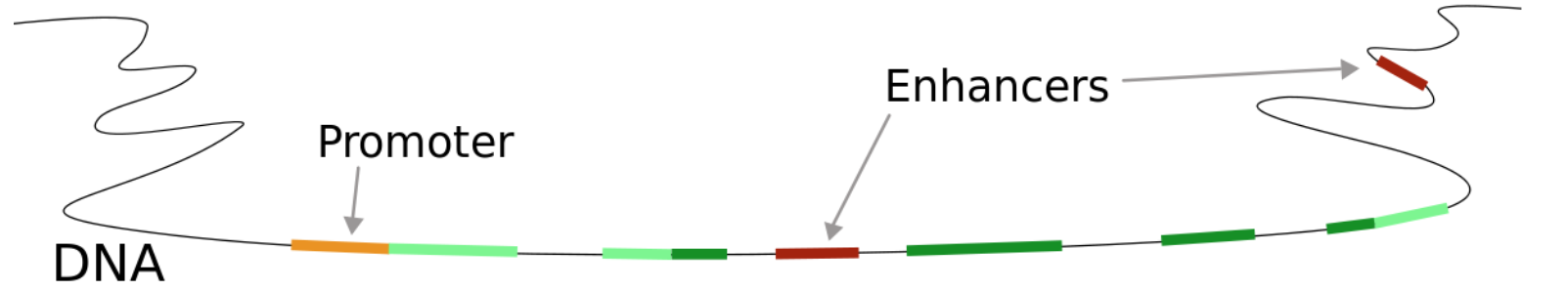
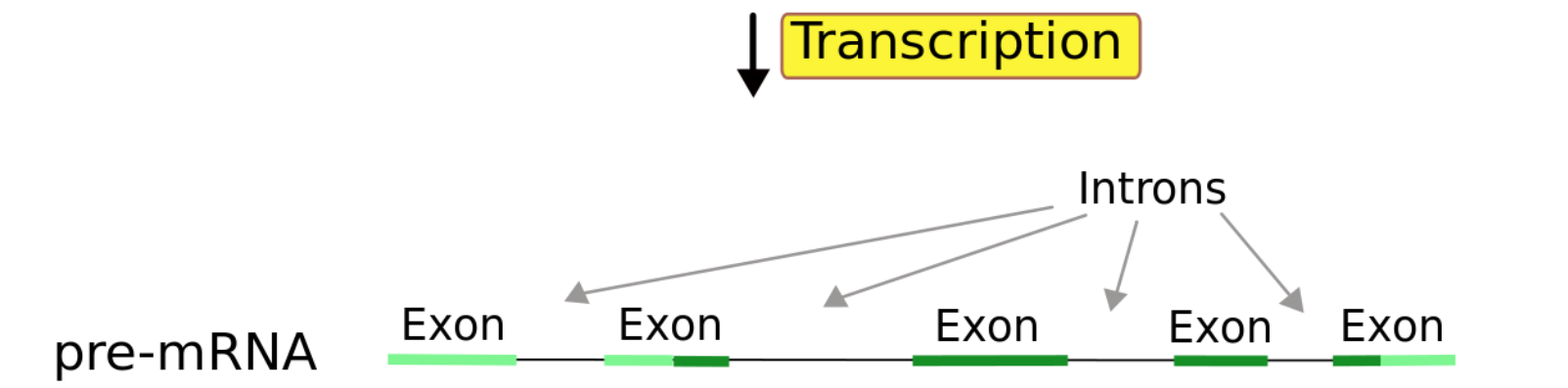
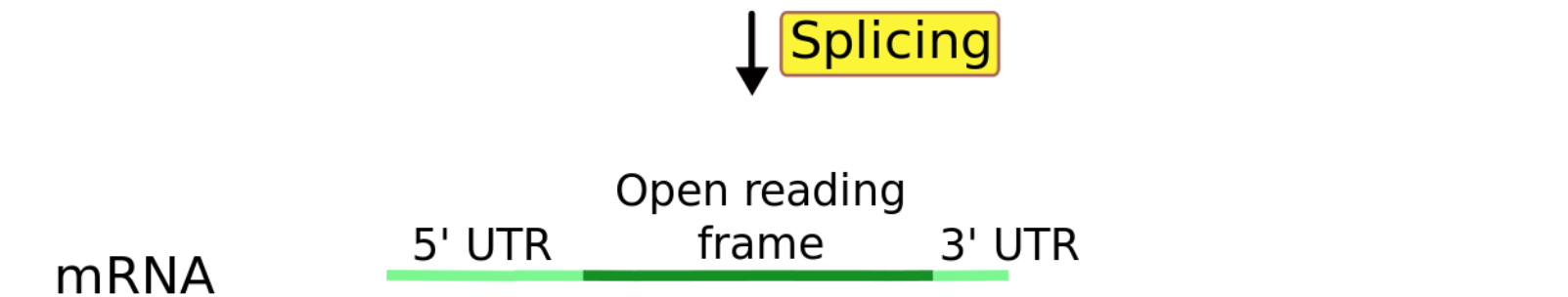
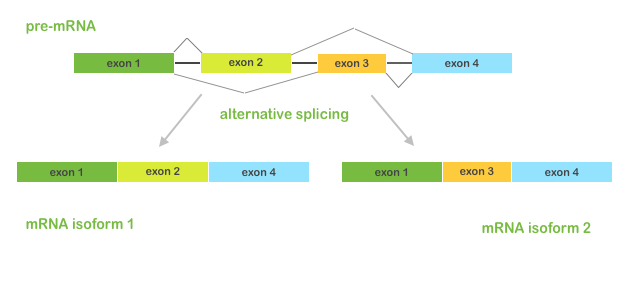
From a single gene, multiple transcripts(isoforms) can (and usually will) be formed.
Why measure gene expression?¶
Because gene expression correlates with protein expression!
Even though nearly every cell in an organism's body contains the same set of genes, only a fraction of these genes are used in any given cell at any given time. It is this carefully controlled pattern of what is called "gene expression" that makes a liver cell different from a muscle cell, and a healthy cell different from a cancer cell.
By measuring gene expression, we can identify active and inactive genes in a cell or tissue. This knowledge is important for drug discovery and creating diagnostic tests.
Before sequencing - library prep!¶
Choosing the appropriate sequencing protocol:
Most of the RNA in a cell is ribosomal RNA (an RNA component of ribosome which is approx 60% rRNA and 40% protein). This is an issue since most scientists (and enthusiasts like us) will be interested in mRNA because of its protein coding function. There are two popular methods for increasing mRNA concentration in your samples:
- poly (A) capture
- ribosomal RNA depletion
- There are also total-RNA protocols that do not enrich for a specific RNA type - for "total RNA" studies;
- Different fragment sizes;
- Bulk or single cell RNA-Seq;
- ...
Then what? Microarrays or RNA-Seq!
DNA microarrays¶
- Around since late 80's.
- Microscope slides with thousands of tiny spots, each spot containing a known DNA sequence or gene. These sequences act as probes to detect gene expression.
- Molecules in the sample are labeled with fluorescent probes.
- The process in which the sample molecules bind to the DNA probes on the slide is called hybridization.
- Following hybridization, the microarray is scanned to measure the expression of each gene printed on the slide.
YouTubeVideo('1_wDrqgS8w8', width=1024, height=576, end = 20)
RNA-Seq¶
- A major breakthrough (replaced microarrays) in the late 00’s and has been widely used since.
- Uses next-generation sequencing (NGS) to reveal the presence and quantity of RNA in a biological sample at a given moment.
- Able to detect novel (undiscovered) isoforms and has a broader dynamic range compapred to microarrays.
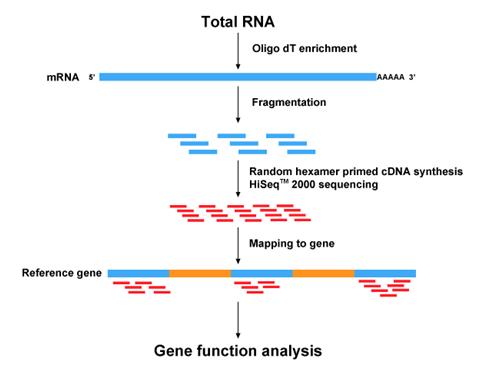
YouTubeVideo('womKfikWlxM', width=1024, height=576)
RNA-Seq analysis goals¶
- Reconstruct the full set of transcripts (isoforms) of genes that were present in the original cells. This catalogue of transcripts is called transcriptome.
- Estimate the expression levels for all transcripts.
The basics will be laid out in following lectures:
- Introduction
- Alignment
- Quantification
- Differential expression